Automatic brain structure segmentation in children with brain tumours
Abigail Bryce-Atkinson,
United Kingdom
PO-1626
Abstract
Automatic brain structure segmentation in children with brain tumours
Authors: Abigail Bryce-Atkinson1, Lydia J Wilson2, Eliana Vasquez Osorio1, Andrew Green1, Gillian Whitfield3, Martin G McCabe1, Thomas E Merchant2, Marcel van Herk1, Austin M Faught2, Marianne C Aznar1
1The University of Manchester, Division of Cancer Sciences, Manchester, United Kingdom; 2St. Jude Children’s Research Hospital, Department of Radiation Oncology, Memphis, USA; 3The University of Manchester, Manchester Academic Health Science Centre, The Christie NHS Foundation Trust, Manchester, United Kingdom
Show Affiliations
Hide Affiliations
Purpose or Objective
Auto-segmentation
tools have been widely implemented in neuroimaging research, enabling extensive
brain segmentations to be obtained with little to no manual interaction. Applying
these tools in paediatric radiotherapy research could enable analyses that
include a wider range of structures than are routinely delineated, be of
benefit for standardising contours in multi-centre studies and allow extensive
dose-effect studies. These tools are developed in adults, so their
applicability in children with cancer is unclear due to age-related differences
and the presence of the tumour and other pathology. This study compares
contours from three auto-segmentation tools in healthy children and in children
with brain tumours.
Material and Methods
We examined
T1-weighted MRIs from 40 healthy children (age 5.0-16.4 years, median 9.3 years)
and 40 children/young adults with brain tumours (including medulloblastoma,
low-grade glioma and astrocytoma; age 1.8-25.2 years, median 8.9 years). Segmentations
of 15 subcortical structures (accumbens, amygdala, caudate, hippocampus,
pallidum, putamen and thalamus bilaterally, and brainstem) were generated by 3
open-source packages: FreeSurfer
v7.2.0, the FMRIB Software Library v6.0.5 FIRST tool (FSL), and the
Computational Anatomy Toolbox v12.8 (CAT). Failed segmentations are reported
but excluded from further analyses. We assessed consistency between each
package via comparison of each structure’s centre-of-mass (CoM), Dice
similarity coefficient (DSC), 95% Hausdorff distance and average contour
distance. We performed ANOVA to evaluate differences between each pairwise software
comparison for each similarity metric, and t-tests to compare differences
between healthy children and children with brain tumours.
Results
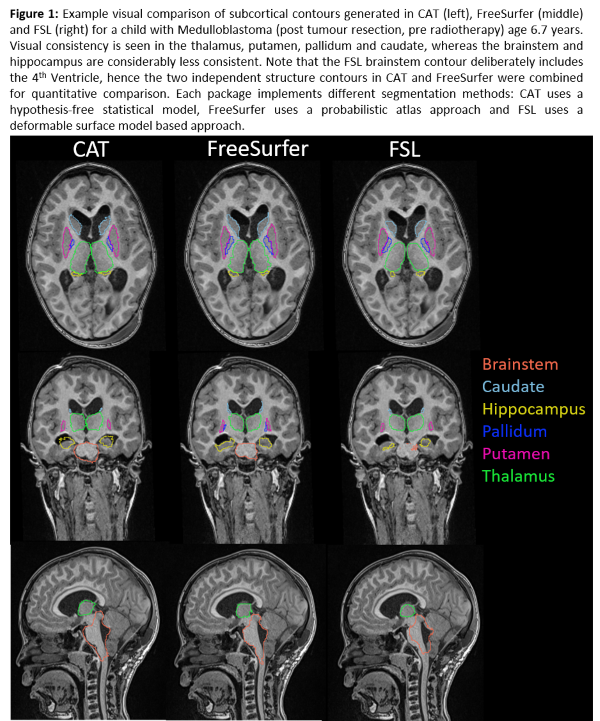
Visual
contour quality was acceptable (Figure 1). Segmentation failed in 11 cases (9
FSL, 1 FreeSurfer, 1 FreeSurfer/FSL), predominantly due to atypical anatomy
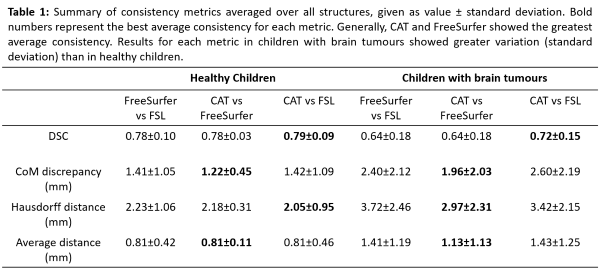
e.g. enlarged ventricles, or poor scan quality. CoM discrepancies and DSC
scores revealed significant differences (p <0.05) between FSL contours and
both CAT and FreeSurfer, but not between CAT and FreeSurfer. FSL contours were
significantly different from FreeSurfer in average distance analyses and from
CAT in Hausdorff distance analyses. We found lower DSC scores, larger CoM and
contour distances, and larger standard deviations within each metric for every structure
in children with brain tumours compared to healthy children. The difference was
significant in analysis considering all structures (Table 1).
Conclusion
The
greater magnitude and variation in similarity metrics in children with brain
tumours suggests auto-segmentation tools perform worse than in healthy children. Contour
differences remained within 4mm in children with brain tumours. FreeSurfer and
CAT were the most consistent and showed the fewest failures, and therefore show
promise for use in paediatric radiotherapy research. Further work validating
against clinical contours is needed.