Self-supervised image feature extraction for outcomes prediction in oropharyngeal cancer
Baoqiang Ma,
The Netherlands
PO-1777
Abstract
Self-supervised image feature extraction for outcomes prediction in oropharyngeal cancer
Authors: Baoqiang Ma1,2, Jiapan Guo1,2,3, Hung Chu1,4, Alessia De Biase1,2, Nikos Sourlos2,5, Wei Tang2,6, Johannes A. Langendijk1, Peter M.A. van Ooijen1,2, Stefan Both1, Nanna M. Sijtsema1
1University of Groningen, University Medical Center Groningen, Department of Radiation Oncology, Groningen, The Netherlands; 2University of Groningen, University Medical Center Groningen, Machine Learning Lab, Data Science Center in Health (DASH), Groningen, The Netherlands; 3University of Groningen, Bernoulli Institute for Mathematics, Computer Science and Artificial Intelligence, Groningen, The Netherlands; 4University of Groningen, Center for Information Technology, Groningen, The Netherlands; 5University of Groningen, University Medical Center Groningen, Department of Radiology, Groningen, The Netherlands; 6University of Groningen, University Medical Center Groningen, Department of Neurology, Groningen, The Netherlands
Show Affiliations
Hide Affiliations
Purpose or Objective
Prognostic outcome models using clinical and
image data make it possible to select the most optimal treatment method for
individual oropharyngeal squamous cell carcinoma (OPSCC) patients. Deep
learning based image feature extraction methods can identify more complex
patterns. The aim is to build Cox
models with the capability of predicting outcomes prior to treatment using
clinical data, image features of CT extracted by an Autoencoder and the
combination of clinical and image features and to compare their performance.
Material and Methods
The OPC-Radiomics set
(https://www.cancerimagingarchive.net/collections/) includes 606 oropharyngeal squamous cell carcinoma (OPSCC) patients.
Pre-treatment planning CT, GTV contours of primary tumours (GTVp) and clinical
parameters were available for 524 patients. The candidate clinical parameters
for building outcome prediction models are gender, age, WHO performance status,
TNM-STAGE, oropharynx cancer stage, treatment modality and HPV status. The
endpoints were local recurrence (LR), regional recurrence (RR), local and
regional recurrence (LRR), distant metastases free survival (MET), tumour-specific
survival (TSS), overall survival (OS) and disease-free survival (DFS). The clinical
parameters were analysed as categorical variables, except that age is
normalized by dividing by 100. Tumour images of 64x64x64 pixels were extracted
around the centre of mass of the GTVp contour. All pixel values outside the GTVp
contour were set to zero.
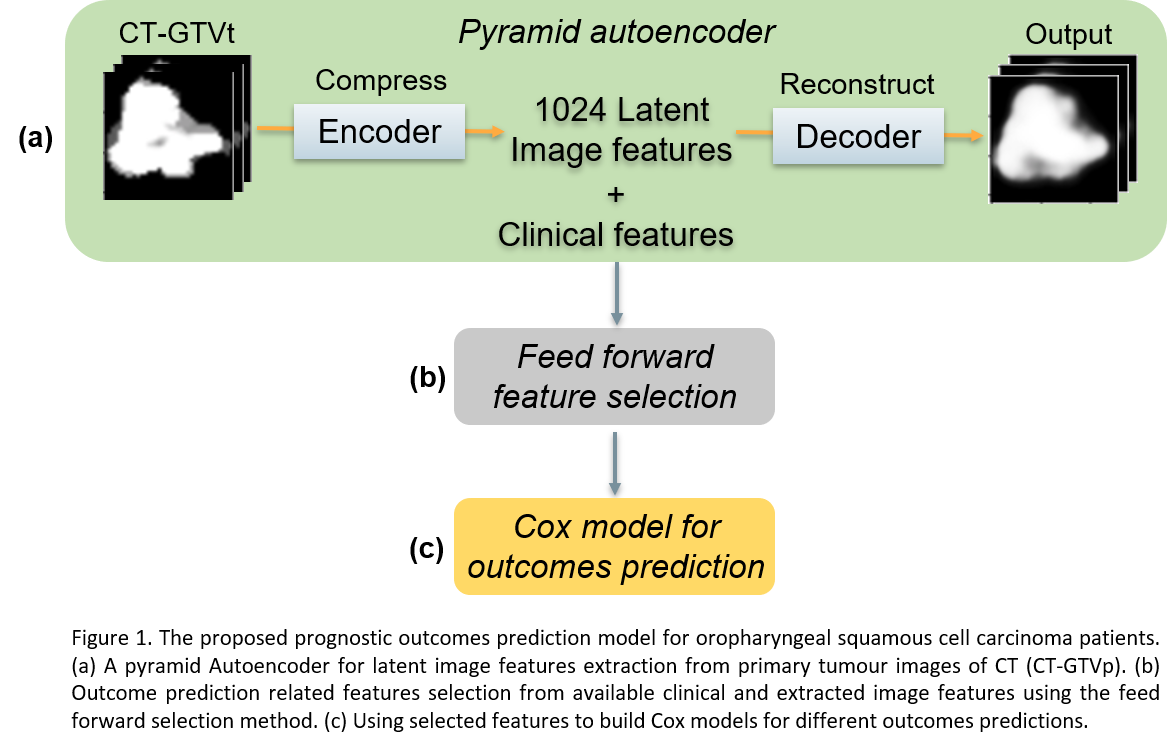
As illustrated in Figure 1, we
propose a deep learning based pipeline to extract image features for outcome
prediction. Firstly, we utilize a 3D Autoencoder for extracting 1024 latent
features that are representative for the tumour images of the cohort. The
Autoencoder architecture is a pyramid Autoencoder for combining different-level
features. The Adam optimizer with initial learning rate 0.001 was used to train
the Autoencoders for 80 epochs. Secondly, for each outcome, a feed forward
feature selection method was applied to select the most predictive features for
the outcome prediction from both extracted image features and clinical
features. Finally, the selected features are used to build Cox models that calculated
the risk score on each outcome for individual patients.
Results
Table 1 displays the C-index results
of the Cox models for each outcome on the training set and test set. The
selected clinical features and the number of selected image features are also
shown here. The Cox models built by clinical and image features together resulted
in higher training C-index values than the models using only clinical or image
features in all seven outcome predictions and higher testing C-index values in
six outcome endpoints except for MET.

Conclusion
Our proposed self-supervised learning based
method which extracts image features of OPSCC patients and uses them in
combination with clinical data shows better predictive performance than the
clinical models for all outcome endpoints.