Combining Single-Organ Deep Learning Segmentation Models for Total Marrow Irradiation
PD-0318
Abstract
Combining Single-Organ Deep Learning Segmentation Models for Total Marrow Irradiation
Authors: Leonardo Crespi1,2, Paolo Roncaglioni1, Damiano Dei3,4, Nicola Lambri3, Pietro Mancosu5, Daniele Loiacono1
1Politecnico di Milano, Dipartimento di Elettronica, Informazione e Bioingegneria, Milan, Italy; 2Human Technopole, Centre for Health Data, Milan, Italy; 3Humanitas University, Department of Biomedical Sciences, Pieve Emanuele, Milan, Italy; 4IRCCS Humanitas Research Hospital, Radiotherapy and Radiosurgery Department, Rozzano, Milan, Italy; 5IRCCS Humanitas Research Hospital, Medical Physics Unit, Radiotherapy and Radiosurgery Department, Rozzano, Milan, Italy
Show Affiliations
Hide Affiliations
Purpose or Objective
Deep Learning (DL) has been successfully applied to the segmentation of organs at risk (OARs). However, when the segmentation task involves many multiple organs, as is the case of the total marrow irradiation (TMI), the application of DL approaches presents additional challenges: (i) a large dataset including all the relevant organs might not be available to train a multi-target DL model; (ii) the combination of many single-organ DL models is not trivial. In this work, we evaluated two different approaches to combine single-target DL models to segment multiple organs and compare them with a multi-organ DL model. This study falls into the ongoing project AuToMI, to automate the TMI planning procedure.
Material and Methods
Several U-Net, DeeplabV3, and Se-ResUnet models were trained on two public datasets: the StructSeg dataset that contains chest CT scans of 50 patients with 6 annotated OARs and SegTHOR that contains CT scans of 60 patients with 4 annotated OARs. The first dataset was split in two subsets: (i) to train the multi-organ and single-organ models and (ii) to evaluate the performance of the two approaches. The second dataset was used only to train some single-organ models for specific OARs, to test our approach on combining models trained from different data sources.
We propose and compare two methods to combine many single-organ models to segment multiple organs: (i) output combination, consisting in applying a 1x1 convolutional layer to combine the output logits of all the single-organ models to compute a multi-organ target; (ii) features combination, consisting in using a convolutional network to combine the features extracted from the last layer of the single-organ models. These methods have been also compared to a multi-organ DL model trained directly from a multi-organ dataset (StructSeg), computing both the Dice Score and the 95th percentile of the Hausdorff distance (HD95).
Results
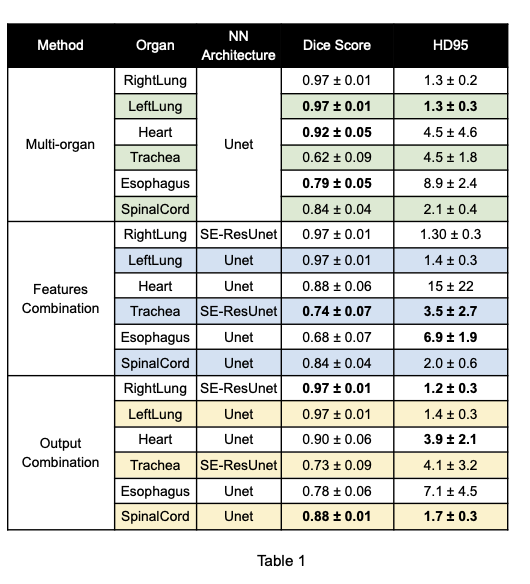
The proposed methods achieved a performance similar or better than the one achieved by the multi-organ model (Table 1). In particular, the proposed methods achieved a better Dice Score on three OARs out of six and a better HD95 in five OARs out of six when compared to the multi-organ model. In particular, the results of HD95 might suggest that combining single-organ models allows to compute a more reliable segmentation.
Conclusion
The proposed methods achieved a performance similar or better than the one achieved by the multi-organ model (Table 1). In particular, the proposed methods achieved a better Dice Score on three OARs out of six and a better HD95 in five OARs out of six when compared to the multi-organ model. In particular, the results of HD95 might suggest that combining single-organ models allows to compute a more reliable segmentation.