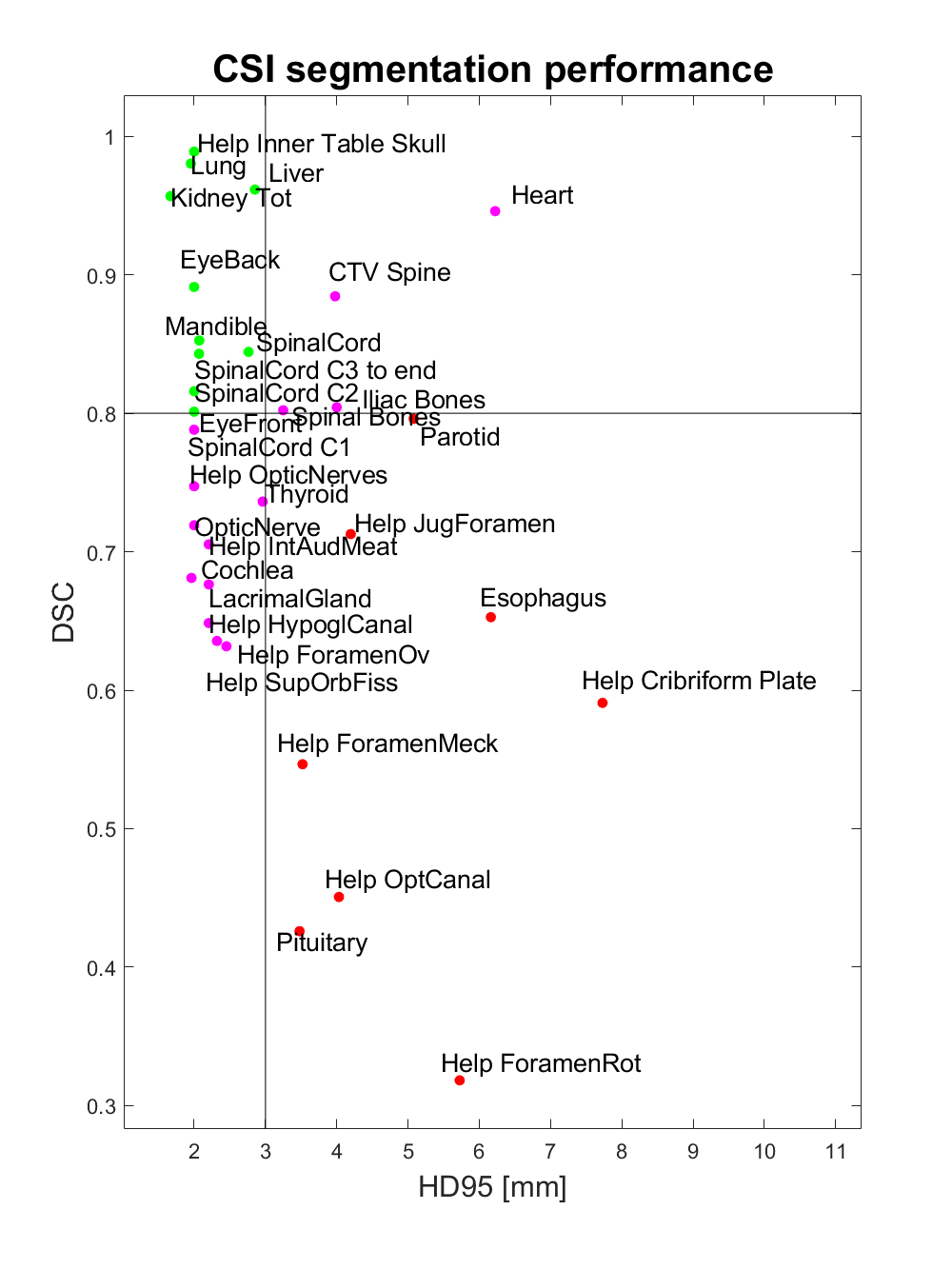
Of the thirty-three structures evaluated, ten structures had a DSC above 0.8 and at the same time a HD95 below 3mm (EyeBack_LR, EyeFront_LR, Kidney_LR, Liver, Mandible, SpinalCord, Lung_LR, SpinalCord_C2, SpinalCord_C3 to end, Help_Inner Table Skull). Eight structures had DSC below 0.8 and HD95 above 3 mm (Parotid_LR, Esophagus, Pituitary, Help_Cribriform Plate, Help_ForamenMeck, Help_ForamenRot, Help_JugForamen, Help_OptCanal). The remaining fifteen structures had either a DSC below 0.8 or a HD95 above 3 mm (Cochlea_LR, LacrimalGland_LR, OpticNerve_LR, Heart, Iliac Bones, Thyroid, Lens_LR, SpinalCord_C1, CTV_Spine, Help_ForamenOv, Help_HypoglCanal, Help_IntAudMeat, Help_OpticNerves, Help_SupOrbFiss, Spinal Boes). (Figure 2)