A clinical evaluation of automatic open-source segmentation algorithm for lung cancer patients
PD-0160
Abstract
A clinical evaluation of automatic open-source segmentation algorithm for lung cancer patients
Authors: Agon Olloni1,2, Carsten Brink3,4, Tine Schytte5,6, Nis Sarup3, Ebbe Laugaard Lorenzen3,4
1Odense University Hospital , Department of Oncology , Odense , Denmark; 2University of Southern Denmark , Institute of Clinical Research , Odense , Denmark; 3Odense University Hospital, Laboratory of Radiation Physics, Odense , Denmark; 4University of Southern Denmark, Institute of Clinical Research, Odense , Denmark; 5Odense University Hospital, Department of Oncology, Odense , Denmark; 6University of Southern Denmark, Institute of Clinical Research, Odense, Denmark
Show Affiliations
Hide Affiliations
Purpose or Objective
Correct delineation of organs at risk (OAR) is essential in thoracic radiotherapy (RT). However, the delineation process can be time-consuming and vary between clinicians. Automatic segmentation is a fast and efficient way of delineating many patients in daily practice and when performing extensive research projects. A deep learning-based segmentation tool, nnU-net, has shown promising results. This study aims to train and validate a nnU-net model for segmentation of left/right lungs, trachea, heart, bronchi, esophagus, and spinal canal.
Material and Methods
We included planning CT-scan and segmentations for all lung cancer patients treated with RT in 2021 at our institution; patients were included if OAR were segmented and no manual segmentations were missing on a slice (n=284). First, 244 patients were used for training a 3D full-resolution U-net in a 5-fold cross-validation approach using the nnU-net framework. The nnU-net was then tested in the latest treated patients (n=40), comparing the manual delineation with the nnU-net segmentation by measuring the dice similarity coefficient (DSC) and the mean surface distance (MSD). Per organ, MSD values were used to define outliers and extreme outliers as those above the 75-percentile value plus 1.5 or 3 times the interquartile range. Next, a medical doctor manually evaluated outliers and extreme outliers with the question, "Would you have corrected the delineations if you were aware of the potential segmentation issue at the time of treatment planning?" Answer possibilities were correct: auto, manual, both, or no correction needed.
Results
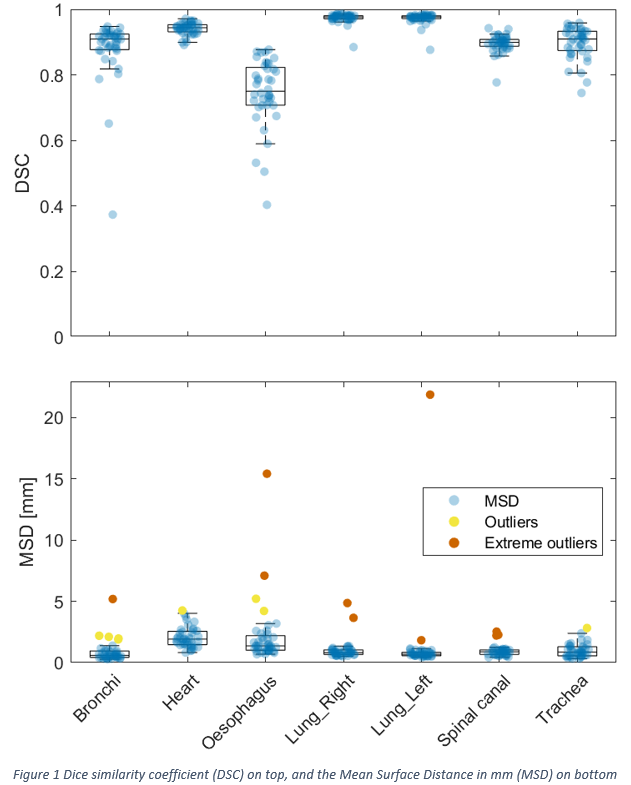
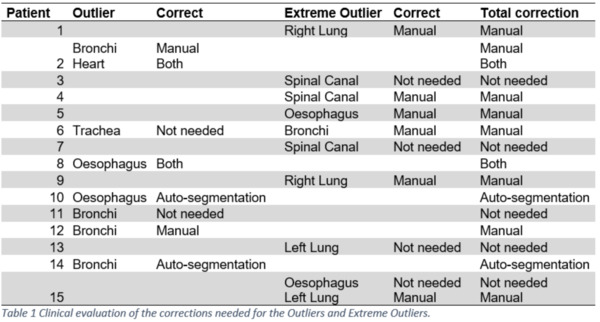
The nnU-net performed well with a DSC of > 0.9 for the heart and lungs. The DSC was lower for the bronchi, spinal canal, trachea, and esophagus, with a DSC of > 0.7 (Figure 1). For all organs, the median MSD was < 1.5 mm. A total of 280 organs were auto-delineated for the final test. Outliers and extreme outliers were identified for 18 organs divided among 15 patients (three patients had outliers in two organs). Of the 18 outliers, 10 of these were scored as extreme outliers. The manual delineation was the problem for all the extreme outliers, as none of the auto delineations required corrections. Among the eight remaining outliers, two automated delineations needed clinically relevant corrections, while the manual and the automated delineations needed corrections for two outliers (table 1). The typical deviation was due to errors in the manual delineation or short manual delineation of long organs such as the esophagus and spinal canal.
Conclusion
The algorithm performed well. Only four of 280 auto-segmentations were categorized as needing corrections in a clinical setting, which was the case for 10 of the clinically used manual delineations. Clinical implementation of the developed algorithm is in process with the hope of reducing delineation errors and the workload of the clinical staff. The trained algorithm will be made freely available for all to use after the publication of the present work.